
About the author: Zoë De Corte (she/her/they/them) has a strong passion for evolution, genomics and bioinformatics. They are a PhD candidate in the lab of Prof. Frederik Hendrickx (University of Ghent & Royal Natural history Museum, Brussels, Belgium) and Prof. Jennifer Brisson (University of Rochester, NY, US). In 2019-2020 they obtained a Fulbright grant and had a 6-month research stay in the Brisson lab.

The recurrent gain and loss of identical traits during the course of evolution has puzzled evolutionary biologists since Darwin. Comprehending the genomic basis of striking cases involving repeated evolution of similar traits is crucial to understand how genetic and developmental mechanisms constrain or facilitate adaptive evolution. In my PhD, I address this evolutionary question by investigating the recurrent evolution of discrete wing dimorphisms across the carabid beetle group.
Source: https://www.coleoptera.org.uk/species/bembidion-properans
Carabid beetles show high diversity in wing morphology. While approximately half of the carabid species are either strictly long-winged or short-winged, a substantial number of species exhibit a remarkable wing dimorphism (wing-dimorphic species) with some individuals developing full wings while others have short wings and can’t fly. Importantly, the evolution of long-winged, short-winged and wing dimorphic species can be found throughout the phylogeny of carabids, which demonstrates that these highly distinct dispersal types evolved repeatedly within this beetle family.
Breeding experiments in carabid beetles revealed that wing development in wing dimorphic species is inherited according to the expectations of a single Mendelian element, with the allele coding for short wings being dominant over the allele coding for long wings. But how could such a simple genetic determination mechanism result in the development of two strikingly different phenotypes?
The objective of my PhD is to find the wing dimorphism locus in one species and then compare that region among other dimorphic species. I used a technique that allowed me to develop markers sampled randomly across the genome (RAD tags) and related the genotypes at these markers with the wing phenotype in the tiny carabid Bembidion properans. Quickly we found a tag that was linked to the wing dimorphism. This felt like a true ‘eureka’ moment, and it seemed that everything had gone smoother than we expected. As this tag represented only a short and not very informative sequence, we sequenced the genome to map the tag. This would allow us to obtain a more complete picture of the locus and the kind of mutations and genes that underlie this dimorphism. I started to reconstruct the genome using short linked reads (Chromium 10x genomics), at that time one of the newer techniques. I had no doubt that I would be able to reconstruct the long and short alleles that gave rise to each morph.
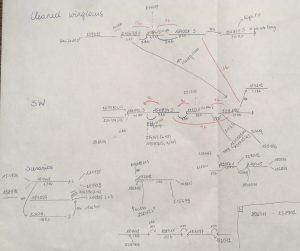
Once I assembled the genome, it became clear that the region was way more complex than I expected. I found several scaffolds linked to the wing differences, with a total length of about 150 kb, but these appeared to be only present in the short-winged allele. I tried to assemble the scaffolds by using all the different kind of available DNA sequence data I had (low coverage pacbio, resequencing data, 10x data & insert size libraries). This ended up with me drawing all my scaffolds by hand, and trying to order and connect them by all possible means I could think of. Looking at these drawings now, I’m really surprised I could still make sense of them (Figure 1). The scaffolds showed many repetitive regions and eventually I realised I was unable to link all of them. It was clear that this assembly would not give us any straightforward answers and that the region is notoriously more complex then I initially assumed.
To continue my quest, I assembled a new genome using a third-generation technique (PacBio) that generates very long reads of 15kb, which would hopefully bridge the repetitive and more complex parts of the genome. This proved to be the key data. The previous 10x scaffolds were assembled in one scaffold, nicely showing that the pacbio reads manage to bridge more complex regions. This new assembly further supported that scaffolds linked to the wings are only present in the short winged-allele and span a genomic region of 200-250kb. I’m currently finalizing the reconstruction of the two alleles.
This journey shows how complex and unexpected the architecture of genomes can be and especially how every genome assembly has its own restrictions depending on the method used. This field is developing so fast. I don’t doubt new techniques will allow us to answer research questions in unprecedented detail.
Once I have fully resolved the two wing alleles, I will annotate the genome to identify the genes localized in this region and those that are differentially expressed between these distinct wing phenotypes. To achieve this, I collected RNA sequences from different developmental stages. I therefore bred B. properans individuals in the lab and collected specimens at three developmental stages: one day after pupation, three days after pupation and at the adult stage. The EECG funding will be used for the transcriptome sequencing. This will not only allow for functional characterization of the wing-dimorphism locus, but result, to our knowledge, in the first annotated carabid genome.



