The genomic bases, or architectures, of complex traits are… complex. But what if by overestimating the complexity of some aspects of the genomic architecture of a trait, we’re actually underestimating the complexity of its evolutionary dynamics? This notion struck me when two things clicked while I was preparing a fellowship application back in 2017.
First, it had become increasingly clear over the last decade or so that a single locus can explain a substantial amount of trait variation, like the Agoutilocus underlying coat colour in deer mice, the optixgene controlling wing patterning in Heliconiusbutterflies, and the RXFP2gene controlling horn architecture in Soay sheep (reviewed in Oomen et al. 2020). What was less clear is how complex traits dominated by a single locus evolve in response to selection, because most of the research to that point was focused on the idea that complex traits were controlled by many loci with only a small effect each. But when Barson et al. (2015) showed that the age at which Atlantic salmon are able to reproduce is largely (approx. 40%) under the control of a single gene (vgll3), Kuparinen & Hutchings (2017) demonstrated that hypothetical single-locus control of this key life-history trait increased phenotypic stochasticity (genetic drift) and led to highly variable and less predictable evolution in response to selection (in this case, fishing) compared to the traditional multi-locus model. This finding led me to rethink how closely coupled the genetic organization of single traits were to emergent population dynamics.
Second, in my own study system of Atlantic cod and many other species (reviewed in Wellenreuther & Bernatchez 2018 and Mérot et al. 2020), it had become clear that structural genomic variation (changes in the abundance, order, or position of DNA sequence) could underlie complex phenotypes and play an important role in adaptation. Structural variants are often characterized by a reduction in recombination between alternative variants. This causes blocks of loci to become trapped in tight linkage disequilibrium. If loci are in complete linkage disequilibrium, then the inheritance pattern of such a ‘haploblock’ resembles that of a single locus.
This raises a fundamental question. Could tightly linked multi-locus architectures show highly variable and unpredictable single-locus evolutionary dynamics like those seen in Kuparinen & Hutchings (2017)? We think they might.
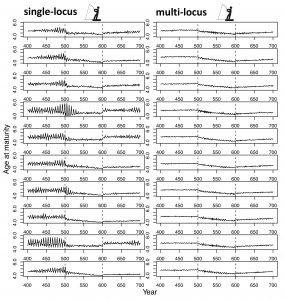
In our new perspective in the Journal of Heredity, we draw from diverse literature to show that eco-evolutionary responses hinge on genomic architecture and that large-effect loci alter evolutionary predictions relative to traditional multi-locus models in a wider variety of situations than shown previously. One of the ways we do this is by extending the Atlantic salmon model (Kuparinen & Hutchings 2017) to a hypothetical trait that doesn’t exhibit sexual dimorphism (like it did in the previous model), to see if we observed similarly unpredictable dynamics. We did. Heightened genetic drift led to nonlinear (i.e., chaotic) dynamics for the evolution of age at maturity in the single-locus but not the multi-locus scenario (Figure 1; Oomen et al. 2020), meaning that these dynamics might be more common than previously considered.
Next, we argue that the inheritance of tightly linked multi-locus architectures resembles that of single loci (Figure 2; Oomen et al. 2020) and go on to show that these architectures are common and probably play a large role in adaptation to rapid environmental change. Finally, we review how linked architectures are favoured by gene flow and underlie diverse traits in natural populations that are directly (e.g., temperature) or indirectly (e.g., biotic interactions) under environmental selection.
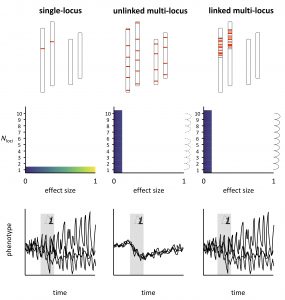
Importantly, one need not dissect the complete genetic architecture of a trait to appreciate that large-effect variants (i.e., single large-effect loci or tightly linked multi-locus architectures) will increase phenotypic stochasticity compared to traditional multi-locus models. The addition of small-effect loci to a large-effect variant (which we refer to as ‘mixed-effect architectures’) would only partially alleviate the uncertainty of the evolutionary response.
When applied to predictions of population responses to environmental change for the purposes of improving conservation, management, and breeding programs, the precautionary principle (Cooney 2004) would dictate considering the chaotic dynamics of the single-locus scenario as a worst case. If, in reality, a mixed-effect architecture underlies the trait, then you might expect an intermediate level of uncertainty according to a recent study by Kardos & Luikart (2020).
Regardless, the traditional multi-locus view of adaptation may lead to a false sense of security regarding the predictability of evolutionary change. In this way, by overestimating the complexity of a trait (e.g., the number of contributing loci), we risk underestimating the complexity (e.g., nonlinearity) of its evolutionary dynamics.
References:

Rebekah Oomen is a James S. McDonnell Foundation Postdoctoral Fellow in Understanding Dynamic and Multi-scale Systems currently working at the Centre for Ecological & Evolutionary Synthesis at the University of Oslo and the Centre for Coastal Research at the University of Agder on understanding how Atlantic cod respond to the environment. She aims to bridge the gap between genomics and eco-evolutionary modelling to better understand how natural populations respond to global anthropogenic stressors, like harvesting and the climate crisis. She completed her MSc and PhD in Ecology and Evolution at Dalhousie University after a bachelor’s degree in Forensic Science at Trent University. Through her work, she strives to mitigate injustice against biodiversity.



