
About the Author: Drew Schield (@drschield) is an NSF postdoctoral fellow working in Dr. Rebecca Safran’s lab at University of Colorado Boulder. He is interested in how evolutionary processes shape genomic diversity and identifying genomic regions that underly adaptive traits and promote reproductive isolation. His work also focuses on the evolution of sex chromosomes and their role in speciation, using barn swallows (Hirundo rustica) and rattlesnakes (genus Crotalus) as model systems.
I’m fascinated by how sex chromosomes evolve, how they differ from other genomic regions, and how these differences may give them special relevance to speciation and adaptation. Along with a group of terrific collaborators, I previously looked into the origins of rattlesnake sex chromosomes, characterized regions bearing signatures of stepwise recombination suppression (i.e., evolutionary strata), and quantified regional dosage compensation on the rattlesnake Z chromosome (Schield et al. 2019). Despite this background, I fell more-or-less backwards into this specific project thanks to a fortunate union of data in-hand, key papers that were fresh on my mind, and new software.
I was a month into my current position when the COVID pandemic started, putting the brakes on some of my plans related to barn swallow speciation genomics (I’m happy to report things are now moving along nicely). In the meantime, I had been reading papers on a commonly observed pattern in nature: sex chromosomes typically harbor low within-population genetic diversity and high between-population genetic differentiation compared to autosomes. There are a number of potential drivers of this phenomenon, including the impact of lower effective sex chromosome population size (i.e., genetic drift), faster fixation of alleles due to low recombination rate and linked selection, and reduced rates of sex-linked gene flow relative to autosomes. Since mutation ultimately provides variation upon which each of these factors can act, mutational biases can also have dramatic impacts on sex-linked patterns (Ellegren 2007). For an excellent overview of these mechanisms, check out Darren Irwin’s (2018) review paper. Each of these factors (and indeed combinations of them) can culminate in a distinctive sex-linked genomic landscape of population differentiation, effectively representing ‘hotspots’ of divergent genotypes, and leading many to hypothesize that sex chromosomes are especially relevant to speciation. What is particularly motivating about this hypothesis is the ability to test for evidence of the roles of these mechanisms by comparing levels of genetic diversity and divergence between sex chromosomes and autosomes.
At this time, I was lucky to have a hard drive full of rattlesnake whole genome resequencing data from my work with Dr. Todd Castoe and access to a beefy analysis server. Inspired by the papers described above, I wanted to dig into my rattlesnake data to test for evidence that various unique features of sex chromosome evolution are relevant to snake speciation. I reasoned that looking for evidence of sex-biased mutation rates, which are present in a wide array of organisms, would be a reasonable first step. Evidence of male-biased mutation rates in snakes had been hinted at previously by Vicoso et al. (2013) based on analyses of coding regions between deeply divergent species. I was keen to confirm this using data from whole genomes from populations, using formulas derived from Miyata et al. (1987) to compare levels of sequence divergence between the Z chromosome and autosomes.
To do this, my co-authors and I measured net nucleotide differences (da) to approximate sequence divergence (d) across the genome between rattlesnake species, which has the benefit of being able to easily leverage population-scale data by comparing within-species nucleotide diversity to between-species absolute differentiation (dxy). The thing is, each of these estimators issensitive to missing data, which can be common in whole genome resequencing datasets. Fortunately, around this same time a pair of savvy population geneticists wrote new software, Pixy, which explicitly accounts for missing genotypes (Korunes and Samuk 2021). We proceeded with anddxy estimation in Pixy, calculated da between species, and plugged these estimates into formulas to derive the ratio of male-to-female mutation rate (αm) and also the ratio of Z chromosome-to-autosomal mutation rate (μΖ/μΑ). Our results, presented in Schield et al. (2021), demonstrate a roughly two-fold male-biased mutation rate in rattlesnakes (Figure 1). As suggested for other organisms, this is a likely consequence of a greater number of germline mutations in spermatogenesis. Using this ratio, we inferred that the Z chromosome mutation rate is roughly 10% higher than autosomes, and while the potential consequences of male-biased mutation are numerous, we are very interested in how this influences patterns of adaptation and reproductive isolation in sex-linked genes, which will be subjects of future work.
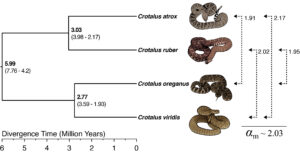
This study presented me with an opportunity to combine my training in population genetics with my ever-expanding interests in sex chromosome evolution in a truly fascinating vertebrate system. I hope it can also serve as an example (adding to a large and growing body of work in other systems) of the interesting genome biology we can uncover using data from sex-linked regions of the genome.
References



