**The AGA grants EECG Research Awards each year to graduate students and post-doctoral researchers who are at a critical point in their research, where additional funds would allow them to conclude their research project and prepare it for publication. EECG awardees also get the opportunity to hone their science communication and write posts over their grant tenure for the AGA Blog. In the first in the series, our EECG awardees write about their research and their interests as an ’embarkation’.**

About the Blog Author: Henry is a PhD candidate at the University of Michigan in the lab of Trisha Wittkopp. He is broadly interested in the evolution of developmental transcription factors and gene regulatory networks more generally.
Nine years ago, I sat down in my Organismal Biology course and opened a notebook with the scarce enthusiasm to be expected of a 19-year-old in an 8 a.m. class. As class time approached, the professor walked to the white board and, in relatively dramatic fashion, wrote the phrase,
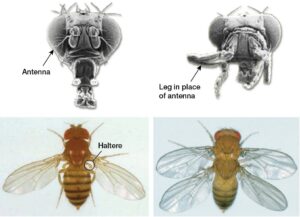
“Evo-Devo”. He turned to face the class and explained that this phrase was shorthand for evolutionary developmental biology, and then proceeded to display a series of images that were both fundamental to the field and disturbing. Most memorable of these images were some Evo-Devo classics – one showing a deformed fly face with legs in place of antennae due to misexpression of the homeobox gene, Antennapedia, and the other in which the fly was given two sets of wings (like a dragonfly) due to a loss-of-function mutation in another homeobox gene, Ubx (see figure). I was in utter awe of these homeobox genes: how could a single gene have such a large influence on development? Perhaps even more stunning, many of these genes played the same developmental role in lineages as distantly related as mammals and insects – how could organisms so different develop through such similar mechanisms? By the end of that lecture, I was overwhelmed by these questions and, although I didn’t know it then, had committed myself to spending the next decade thinking about them.
After many technical and conceptual roadblocks at the start of my PhD, my biggest breakthrough was undoubtedly when I remembered this day nine years ago. Up to that point, my research had broadly focused on the role of chromatin in gene expression evolution, and through this work I found myself drawn to this class of molecules known as pioneer factors – transcription factors with the unique ability to bind inaccessible DNA and “pioneer” the regions for subsequent activators to bind (Larson et al. 2021). After struggling for years to articulate my interest in these molecules, I found clarity through my recollection of my initial excitement at homeobox genes. Many pioneer factors are like homeobox genes (and some homeobox genes are in fact pioneer factors) in both a functional and evolutionary sense: they can reverse, direct, and reprogram cellular differentiation and some are deeply conserved over hundreds of millions of years (Larson et al. 2021). Through this connection, I realized that pioneer factors excited me because they provided the mechanistic explanation I had yearned for with the early Evo-Devo findings.
The direction of my thesis was now clear: I wanted to understand if, when, and how pioneer factor molecules evolved. For example, is the mechanism by which these pioneer factors remodel chromatin through development also conserved across hundreds of millions of years, or does the mechanism evolve as well? I have chosen to focus on the specific pioneer factor, Grainyhead (Grh), because 1) Grh orthologs as distantly related as fly and mouse similarly drive epithelial cell differentiation (Kim & McGinnis, 2010 & Ting et al. 2005) and 2) there is a rich literature in which multiple functional regions – DNA binding, dimerization, pioneering, and transactivation – within the protein have been mapped (Attardi & Tjian, 1993). The project supported by funds from the EECG grant will untangle which of these Grh functions have or have not evolved, which will shed light on the changing and unchanging parts of epithelial development across metazoans.
References



