**The AGA grants EECG Research Awards each year to graduate and post-doctoral researchers who are at a critical point in their research, where additional funds would allow them to conclude their research project and prepare it for publication. EECG awardees also get the opportunity to hone their science communication and write three posts over their grant tenure for the AGA Blog. In the first in the series, our EECG awardees write about their research and their interests as an ’embarkation’.

About the author: Jeremy is an evolutionary ecologist and NSF postdoctoral fellow working with Catherine Linnen at the University of Kentucky Department of Biology. He is broadly interested in adaptation, speciation, population genomics and chemical ecology of insects. Follow him on Twitter @flydrocarbon.
Biological introductions provide unique opportunities as ‘natural experiments’ to gain insights into the process of adaptive shifts to novel hosts. A long-standing paradox in invasion biology asks: given the profound reduction in genetic variation that accompanies many species introductions, how do invading populations adapt (Frankham 2005)? Despite an abundance of natural examples and relevance for addressing this paradox, relatively few studies have investigated the adaptation of introduced insects to native hosts.

In Connecticut in 1914, the invasive pine sawfly Diprion similis was first observed in North America, thereafter spreading across the eastern half of the continent in conjunction with a noticeable shift in host (Figure 1). While in its native range in Europe, D. similis is found on thick needled pine species, North American D. similis primarily attacks white pine (P. strobus), which has exceptionally thin needles compared to other conifers. Because ovipositing females must embed their eggs within the needle without cutting too deeply, thin needles can represent a substantial barrier to host shifts (Bendall et al. 2017), and only a single native diprionid—Neodiprion pinetum—feeds primarily on P. strobus.

In a broader set of projects, I have been utilizing both modern and preserved museum collection samples of D. similis dating back to the original introduction to investigate the patterns, modes, and predictability of this recent and stark host shift (Figure 2). As part of this work, Canadian historical host-use records revealed that D. similis did not make a large-scale shift to using white pine until the 1960s, at which point there was a rapid shift to ~90% white pine use within 10 years (Figure 3). This delay in host-shift post-introduction could mean that genetic variation necessary for adaptation to a novel host was not present until this point in time and could be indicative of a second invasion that introduced variants responsible for this rapid host-shift. Multiple introductions have often been proposed as an important or even necessary step in adaptation to novel habitats during biological invasions (Rius and Darling 2014). Demographic models using population genomic data are effective tools for estimating the number of past invasions—provided that the invasion was recent, and sampling is broad enough.
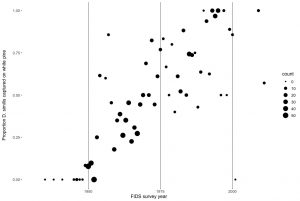
Using EECG 2021 award funding, I aim to test the hypothesis that multiple invasions are needed to provide the necessary genetic variation to fuel D. similis adaptation to white pine in the introduced range. To do so, I will be obtaining low-coverage whole genome sequencing data from a set of 96 geographically diverse D. similis samples. I will then use these data for population genomic and demographic modeling analyses aimed at uncovering the historical demographics of this host shift. This system is particularly suited for evaluating recent and rapid host-shifts, and inferences from these analyses can have sweeping implications on our understanding of the patterns and modes adaptive evolution and host-use diversification in biological invasions.
References
Frankham R (2005) Resolving the genetic paradox in invasive species. Heredity, 94, 385.



