
About the Blog Author:
Colby Behrens is a PhD candidate at the University of Illinois Urbana-Champaign in the lab of Dr. Alison Bell. Colby is interested in the evolution of behaviors and the underlying genetic mechanisms that regulate them. He uses three-spined stickleback fish to explore these patterns.
Animals exhibit tremendous diversity in their social behaviors, especially in reproductive strategies. Despite increasing interest in this topic, the mechanisms that generate this diversity are often poorly understood, in part because the underlying genetic architecture of most behaviors is likely to be complex (Boyle et al. 2017) and because behaviors are often difficult to phenotype in a high throughput manner. Understanding the underlying genetic mechanisms of behavioral divergence could shed light on the evolution and regulation behaviors and other complex traits.

Our lab uses three-spined stickleback (Gasterosteus aculeatus) to investigate these questions. Stickleback are well-suited to laboratory phenotyping, exhibit tremendous natural diversity, and have well-described reproductive behaviors (Wootton 1976). Typically, stickleback fathers are the sole providers of parental care. In most populations, males guard a territory, build a nest, court females (Figure 1), then care for the fertilized eggs. However, males from a unique Nova Scotian population provide almost no care to their offspring. Instead, they remove most or all of the embryos from their nest shortly after fertilization (Blouw 1990). Once the embryos are removed males resume normal courtship activities and do not return to care for their offspring. Population genomic analyses suggest that the non-caring population has recently diverged from the caring population (Samuk 2016) and that the behavioral differences are heritable (Blouw 1996). In addition to parenting differences, my lab has shown that the two populations differ in an entire suite of reproductive behaviors, including nest architecture and courtship behaviors. The non-caring population builds relatively simple nests and engages in high levels of female-directed courtship, while the caring population constructs more elaborate nests and engages in high levels of nest-directed courtship. However, the mechanisms driving this behavioral divergence are still unknown.
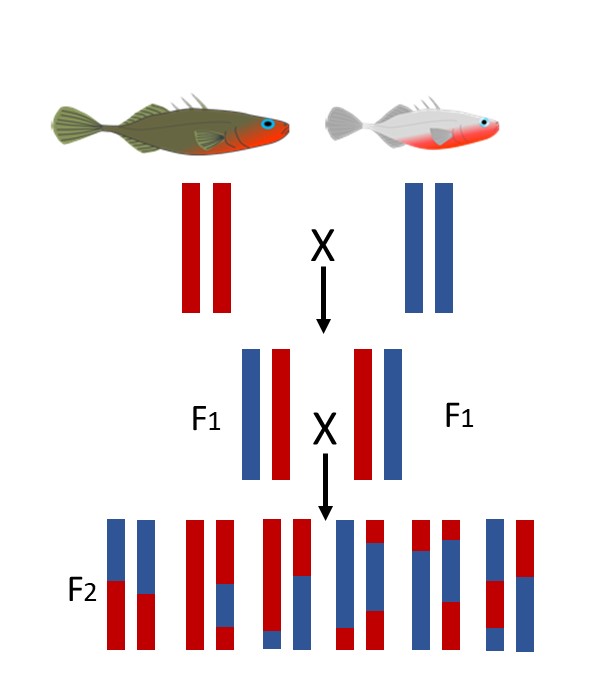
My dissertation seeks to address this question and investigate the mechanisms underlying this reproductive divergence. One way to approach this question is through quantitative trait locus (QTL) mapping (Figure 2), which associates phenotypic variation with regions of the genome by taking advantage of natural recombination rates (Broman 2001).. As a first step, I generated F2 hybrids between the two populations and quantified differences in their territorial aggression, nest architecture, courtship behavior, and parenting behavior in a large mapping population. These hybrids exhibited significant variation in all phenotypes, which is ideal for QTL mapping. Additionally, traits were uncoupled in the F2’s, suggesting that traits will map to different genomic regions.
The AGA EECG award is funding the critical sequencing costs of the project, which will allow me to map divergent behaviors to regions of the genome. The results from the QTL mapping project will then be synthesized with other genomic datasets, including brain RNA expression, to better understand the mechanisms by which divergent reproductive strategies evolve.
References
Boyle, E. A., Li, Y. I., & Pritchard, J. K. (2017). An expanded view of complex traits: from polygenic to omnigenic. Cell, 169(7), 1177-1186.
Wootton, R. J. (1976). The biology of the sticklebacks. United Kingdom: Academic Press.
Blouw, D. M., & Hagen, D. W. (1990). Breeding ecology and evidence of reproductive isolation of a widespread stickleback fish (Gasterosteidae) in Nova Scotia, Canada. Biological Journal of the Linnean Society, 39(3), 195-217.
Samuk, K. M. (2016). The evolutionary genomics of adaptation and speciation in the threespine stickleback (Doctoral dissertation, University of British Columbia).
Blouw, D. M. (1996). Evolution of offspring desertion in a stickleback fish. Ecoscience, 3(1), 18-24.
Broman, K. W. (2001). Review of statistical methods for QTL mapping in experimental crosses. Lab animal, 30(7), 44-52.



