Say you are studying a population of anadromous fish. These fish live in the ocean and swim upriver to spawn, with each fish returning to the river in which it hatched. The different rivers wind up containing separate subpopulations, as fish from different rivers never interbreed. How can these distinct subpopulations be recognized when the fish are in the ocean?
Attaching identification tags quickly becomes unwieldy and expensive. Failing to distinguish between these populations and identify fish stocks correctly can lead to overfishing of subpopulations.
Fisheries researchers have a clever way to solve this problem: identifying parasites. In this situation, a parasite found in only one river serves as a natural tag. Studies have found that using parasite tags can double the chances of correct classification, which allows fisheries managers to make informed choices about how to manage their stocks (Poulin and Kamiya 2013). Parasites can mark different subpopulations or indicate when a population is homogeneous. A study short-finned squid found that squid that were geographically far apart shared a strain of a nematode parasite Anisakis simplexB, showing that these squid were interbreeding (Pascual and Hochberg 1996).
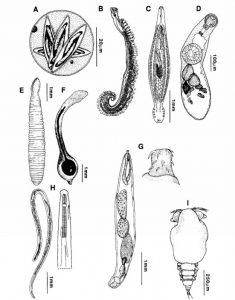
Research on parasite tagging of fish stocks dates back to the 1930s; by the 1980s, genetic identification methods had replaced morphological identification. Genetic identification allows for rapid sampling of large data sets: next-generation sequencing is a convenient way for fisheries managers to sequence large populations quickly and inexpensively (Catalano et al. 2014).
Parasites function as part of the holobiont: the collective genome made up of both a host organism and the symbiotic organisms associated with it (Hedayat and Lapraz 2019). Nuclear DNA from the host provides a lot of information about population structures, but it contains little information about environmental factors and does not always show subtle lineage differences, such the distinction between non-interbreeding populations within a species. The holobiont can create a more complete picture of an animal’s life. Many holobiont studies focus on the gut microbiome, which can provide information about diet and environmental factors such as geographic location, salinity, and water quality (Tarnecki et al. 2017). Parasites can yield similar information and are often easier to collect and identify.
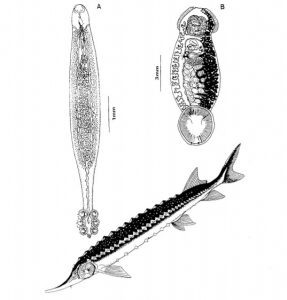
Parasites can be internal or external, large or microscopic, specific to one host or general, simple or complex. Some parasites live exclusively on one host for the duration of their lifecycle, and some rely on complicated multi-host life cycles to mature and reproduce. Single-host parasites are easier to use for identification and data collection, while parasites with multiple-host lifecycles indicate linkages between species. If a seabird is found to contain a parasite found only in one population of fish, researchers can conclude that this seabird has eaten fish from this population.
Parasites come in all shapes and sizes, but a good parasite identification tag needs a few specific characteristics (Williams, MacKenzie, and McCarthy 1992):
- High infection levels: the parasite must be widespread in the host population.
- High host specificity: the parasite must be picky about its choice of host.
- High genetic variability: the parasite must have identifiable genetic variations that can be used to distinguish between subpopulations.
- Long lifespan and stable infection: the parasite must remain committed to staying attached to its host for enough time to conduct population studies.
- Easy to sample: the parasite must be accessible to researchers.
- Minimally harmful: the parasite must not have seriously deleterious effects on host behavior or health.
This sampling method has potential for broad applications in ecological research. Parasite tagging is especially useful in studying species where traditional capture-recapture studies may be impossible, such as deep-sea animals (Catalano et al. 2014). Collecting data on marine species presents unique difficulties: movement patterns in the ocean are difficult to quantify and dead bodies of marine animals usually sink to the ocean floor. Collecting external parasites or fecal samples is less invasive than many other sampling methods that rely on collecting tissue samples, which facilitates sampling of protected wildlife. Including parasites as part of holobiont data set has the potential to expand the boundaries of population datasets, giving ecology researchers new tools to understand cryptic species and complicated community structures.
References

Anne Christian is a conservation biologist and a master’s grad from Tufts University. She explores how population dynamics are altered by cryptic species interactions. Past research has focused on the parasite loading of Atlantic sturgeon in the Hudson River. Follow Anne on Twitter at @AnneChristian94



