
About the Author: Hayden Waller is a PhD Candidate in the Department of Neurobiology and Behavior at Cornell University. He works with Hawaiian crickets to better understand the genetic basis of behavioral variation. You can find him on twitter @tbh_i_dont_know
Next time you find yourself at a party, walk up to a group of strangers and ask how many of them have ever heard of CRISPR/Cas9. Two things will happen. First, you will learn that about half of them will have heard of it. Second, they will probably avoid you the rest of the night. On the one hand, you will be left standing in the corner sipping your drink alone, but on the other, you will be able to remark upon the fact that if you had asked the same question perhaps even as little as five years ago, you would be lucky if one of them said yes. If you repeat the same behavior with a group of life science researchers, not only will all of them have heard of the technology, but at least one will be actively using it, and they will be excited to talk about it. The age of CRISPR has arrived.
The vast majority of news stories discussing the technology are centered around one of two things: sensationalist pieces centered around using CRISPR in humans, or stories that center around its medical applications. This makes sense. Humans tend to be interested in other humans. And to be sure, the promise of CRISPR in medicine has the most potential to directly affect the quality of human life (lest you think this is not already happening, in June of this year a study was published by researchers at the University of Pennsylvania which demonstrated that injecting CRISPR into the blood of patients suffering from a rare but fatal nerve/heart disease shuts off production of a toxic protein by silencing the underlying mutated gene). What is very rarely covered by news outlets is CRISPR’s impact on basic life history research. This is a shame, since it has single-handedly ushered in a New Golden Age of Evolutionary Biology.
To do my part in helping to remedy this attention deficit, I sat down with Dr. Rick Fandino in the Department of Ecology and Evolutionary Biology at Cornell University to discuss the state of the field and where it is heading in the future. He is currently working with Dr. Bob Reed, using CRISPR to understand the regulatory mechanisms of seasonal plasticity in Buckeye butterflies. “What I’m trying to understand is how environmental signals are transduced to genome-wide shifts in expression of pigment genes” he told me. And this is not his first rodeo.
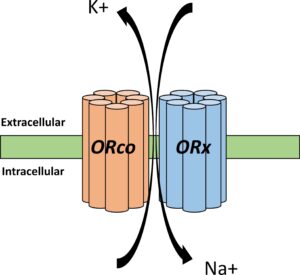
In his previous postdoc at the Max Planck Institute for Chemical Ecology in Jena [Germany] Rick led a project whose goal was to use CRISPR to knockout a gene called orco hawkmoths. orco is a highly conserved gene that forms a heteromeric structure with one of hundreds of ligand-selective odorant receptors (ORs) and is essential for proper olfactory function in insects. The project was a resounding success. Rick and his team were able to show that knocking out orco significantly disrupted a moth’s flight patterns, feeding, and oviposition behavior by reducing their ability to detect olfactory information. Indeed, several other studies have investigated the effects of orco knockouts as well and the findings appear similar across insect orders. If you knockout orco, you get a wide array of behavioral, neurological, and physiological effects. This is, frankly, a stunning revelation about how upwards of three quarters of the animals on planet Earth function and it also serves as an illustrative example of how CRISPR/Cas9 has been used in the field of evolutionary biology for the last five to ten years.
Until recently, the field has been focused on these sorts of upstream, master-regulatory genes (for other examples see fruitless, cortex, Ptgfr). Using CRISPR/Cas9 to knock out these types of genest has given us unprecedented insight into the importance and function of this many very important genetic/behavioral/physiological pathways. I am a huge advocate for continuing to give attention to these types of genes. Master regulators are obviously very important to understand. But let us consider for a moment what we may be missing.
Theory has led us to suspect for decades that there is a negative relationship between mutations in highly pleiotropic genes and an organism’s fitness. In fact, much recent work has been done to test this prediction experimentally (see here for review). Since most mutations are deleterious, a mutation in a highly pleiotropic master regulator is bound to have far-reaching and potentially organism-wide negative consequences. Even if a mutation positively affects a subset of downstream genes, it will still likely affect others negatively. It is therefore exceedingly rare that mutations arising in such genes will carry a net benefit on an organism’s fitness. Therefore, highly pleiotropic genes tend to remain highly conserved in sequence and function across large taxonomic distances. So, while investigating master regulators is useful for learning about the genetic features that make organisms similar to each other, it is the less-pleiotropic downstream genes that will allow us to discover the genetic features that underlie divergence. This is the future of CRISPR in evolutionary biology.
“We can begin to narrow our focus in on genes that underlie very specific behaviors.” said Rick when I asked him about the future. “For example, certain receptors tuned to particular plant or floral compounds.” In other words, if we return to our insect odorant receptor example from before, now that we understand the function of the highly conserved and pleiotropic component (orco), it is time to begin to investigate the ligand-specific, low-pleiotropy components that are more susceptible to directional or divergent selection and thus more likely to drive or maintain species differences. It is not difficult to imagine a scenario in which a mutation in an OR gene results in a host shift and perhaps even a subsequent speciation event. While orco is essential for all odorant receptors to function, it is this ligand-specific OR that underlies this more fine-scale behavior of whether to oviposit/eat plant species A versus plant species B.
I asked Rick what he thought was driving the field forward these days and he said, “CRISPR can’t function without having a better understanding of an organism’s genome and as the price of sequencing drops dramatically, this makes it so average labs can begin to take advantage of the technology.” This, he said, combined with the appearance of new sequencing technologies such as ATACseq that allow us to look at where chromatin is opening and closing, represent the indirect advances that are allowing for the increase in the adoption of CRISPR as an essential tool in the evolutionary biologist’s toolkit.
CRISPR/Cas9 has already changed the world in many ways. For evolutionary biologists, it represents an indispensable tool in our attempts to understand the organisms that populate said world. Much of our attention has been focused on highly pleiotropic master regulatory genes (which ought to continue, by the way) but the time has come to begin investigating the downstream, lowly pleiotropic genes that drive divergence. I know I’m probably not the first one to make the claim that we have entered the New Golden Age of Evolutionary Biology, but I will make it nonetheless. It is certainly an exciting time to study life.
References



