
About the Author
Dr. Chase H. Smith is an evolutionary biologist and a leader in understanding aspects of the biology, ecology, evolution, and conservation of freshwater mussels. Currently he is a research fellow at the University of Texas at Austin mentored by Drs. Justin Havird, David Hillis, and Mark Kirkpatrick where he researches, among many other topics, sex determination in bivalves. For updates on current and future research, follow him on Twitter @Texas_Mussels, Google Scholar, and/or ResearchGate.
When we think about mitochondrial DNA, we often just refer to it as “the powerhouse of the cell” that we get from our mothers. However, as with many other things in science, mitochondrial biology is MUCH more complex. As a malacologist, I have always been interested in a unique mitochondrial biology known as doubly uniparental inheritance (DUI), a phenomenon involving the independent transmission of female and male mitochondria (Fig 1). This biology is unique to bivalves and was originally discovered in the early 1990’s (Hoeh et al. 1991). It has now been revealed in 5 bivalve orders (Gusman et al. 2016; Capt et al. 2020), including my primary study system—freshwater mussels in the order Unionida. Origination hypotheses for the origin of DUI, particularly the origin of male-trasmitted mitochondrial DNA, has remained elusive (Stewart et al. 2009). Many of these questions had come from some preconceived notions of mitochondrial DNA, primarily the fact that it is often considered to not recombine (Boore, 1999), despite the overwhelming growing body of literature challenging this generalization (Barr et al. 2005).
As with seemingly many projects, the origins of my recently published study in Journal of Heredity (Smith et al. 2023; https://doi.org/10.1093/jhered/esad004) began with a discussion in a lab meeting. While reviewing a draft of a now published paper from the Havird lab (Maeda et al. 2021), the lab had a thorough conversation about origination hypotheses of male-transmitted mitochondrial DNA, with a concentration on if it had originated once or many times (up to 10) across bivalves. Primarily focusing on phylogenetics during my time as a graduate student, I found it difficult to “accept” that this complex and seemingly unique trait would have evolved multiple times. This led to a discussion of how you could reject multiple origin hypotheses, especially given the known issues with previous studies in ancestral reconstruction of the trait, primarily due to the known recombination of female- and male-transmitted mitochondrial DNA in some bivalve lineages. Although not needing another side project, as most academics would, I begrudgingly accepted this challenge.
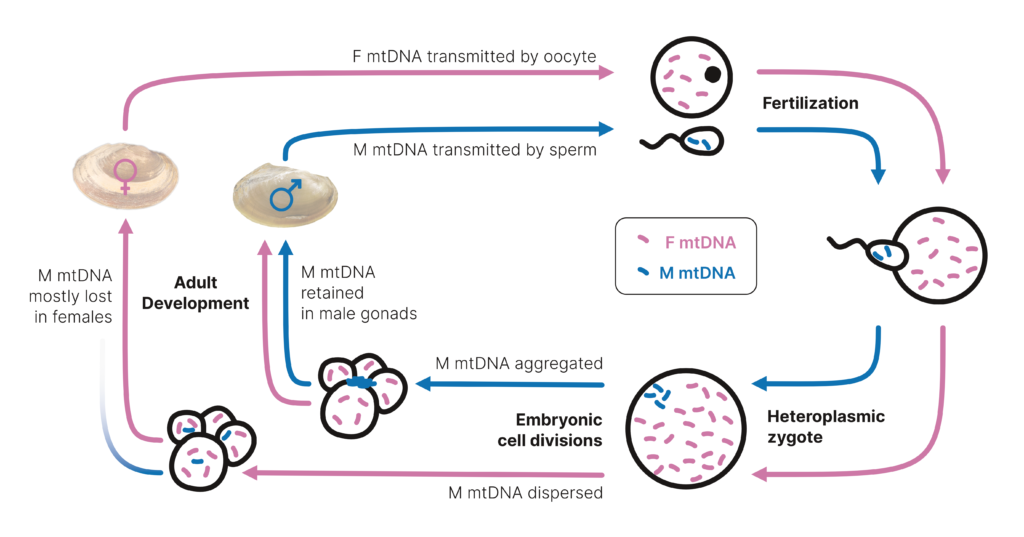
Given traditional ancestral character reconstruction (e.g., Dollo parsimony) could not resolve this issue, recent advancements in site-based methodology seemed like a logical next step to test origination hypotheses. The logic behind this was that the methodology, in theory, would be able to address issues with period recombination occurring between female- and male-transmitted mitochondrial DNA. Specifically, even in the presence of multiple recombination events, phylogenetic signal for a single origin could be retained at individual sites in the male-transmitted mitochondrial DNA that would be masked when using traditional concatenation-based methodology. This thinking was certainly influenced by a recent article (at the time) from Mark Hibbins investigating hemiplasy (Hibbins et al. 2020; https://github.com/mhibbins/HeIST). In our paper, we used a site-based phylogenetic modeling approach to test origination hypotheses of male-transmitted mitochondrial DNA, as well as infer the prevalence of mitochondrial recombination in bivalves. Our results were able to provide statistical support for a single origin of male-transmitted mitochondrial DNA, even in the presence of a complex evolutionary history that included multiple recombination events with female-transmitted mtDNA (i.e. a pattern of concerted evolution). This aligns with previous authors who have hypothesized that male-transmitted mitochondrial DNA originated once and was subsequently lost across many bivalve lineages.
Although our results from empirical data were able to provide insights into an issue that has been argued for nearly 30 years, there is still little known about the role of male-transmitted mitochondrial DNA in biological processes. Specifically, the role of mitochondrial DNA in sex determination or sexual development in bivalves is apparent, given traits of these genomes align with sexual transitions (at least in freshwater mussels; Breton et al. 2011). Given sex determining pathways remain inconclusive in bivalves, continued research into these organisms will continue to provide novel insights into the natural world.
References
Barr CM, Neiman M, Taylor DR. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 2005;168(1):39–50. https://nph.onlinelibrary.wiley.com/doi/10.1111/j.1469-8137.2005.01492.x
Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27(8):1767–1780. https://academic.oup.com/nar/article/27/8/1767/2847916
Breton S, Stewart DT, Shepardson S, Trdan RJ, Bogan AE, Chapman EG, Ruminas AJ, Piontkivska H, Hoeh WR. Novel protein genes in animal mtDNA: a new sex determination system in freshwater mussels (Bivalvia: Unionoida)? Mol Biol Evol. 2011;28(5):16451659. https://academic.oup.com/mbe/article/28/5/1645/1269144
Capt C, Bouvet K, Guerra D, Robicheau BM, Stewart DT, Pante E, Breton S. Unorthodox features in two venerid bivalves with doubly uniparental inheritance of mitochondria. Sci Rep. 2020;10(1):1087. https://www.nature.com/articles/s41598-020-57975-y
Gusman A, Lecomte S, Stewart DT, Passamonti M, Breton S. Pursuing the quest for better understanding the taxonomic distribution of the system of doubly uniparental inheritance of mtDNA. PeerJ. 2016;4:e2760. https://peerj.com/articles/2760/
Hibbins MS, Gibson MJ, Hahn MW. Determining the probability of hemiplasy in the presence of incomplete lineage sorting and introgression. eLife. 2020;9:e63753. https://elifesciences.org/articles/63753
Hoeh WR, Blakley KH, Brown WM. Heteroplasmy suggests limited biparental inheritance of Mytilus mitochondrial DNA. Science. 1991;251(5000):1488–1490. https://www.science.org/doi/10.1126/science.1672472
Maeda G, Iannello M, McConie HJ, Ghiselli F, Havird JC. Relaxed selection on male mitochondrial genes in DUI bivalves eases the need for mitonuclear coevolution. J Evol Biol. 2021;34(11):1722–1736. https://onlinelibrary.wiley.com/doi/10.1111/jeb.13931
Smith CH, Pinto BJ, Kirkpatrick M, Hillis DM, Pfeiffer JM, Havird JC. A tale of two paths: The evolution of mitochondrial recombination in bivalves with doubly uniparental inheritance. J Hered. 2023;esad004. https://academic.oup.com/jhered/advance-article/doi/10.1093/jhered/esad004/7075777
Stewart DT, Breton S, Blier PU, Hoeh WR. Masculinization events and doubly uniparental inheritance of mitochondrial DNA: a model for understanding the evolutionary dynamics of gender-associated mtDNA in mussels. In: Pontarotti P, editor. Evolutionary biology from concept to application II. Berlin (Germany): Springer-Verlag; 2009. p. 163–173. https://link.springer.com/chapter/10.1007/978-3-642-00952-5_9



