Figuring out evolutionary relationships between species is hard enough when they diversified recently, but what if they rapidly diversified many millions of years ago?

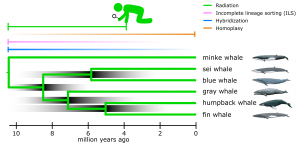
A group of baleen whales, the rorquals (Balaenopteridae), for example, diversified starting about 10.5 million years ago (Figure 1; Árnason et al. 2018). Within this group, the evolutionary relationship of the famous gray whale has long been inconclusive. Traditionally, based on morphological analysis, gray whales were placed in a separate genus that is sister to the rorquals (Nowak 1999). This was done based on a set of characteristics that are unique and different from the other rorquals, such as a dorsal hump followed by a series of 6-12 knuckles instead of a dorsal fin. Only more recently, molecular studies have suggested that they evolved from within rorquals (e.g. Árnason et al. 2018). Yet, depending on the molecular marker used, the exact relationship to the other rorquals can show different results (Árnason et al. 2004; Árnason et al. 2018).
The short period in which they diverged has left genetic signals to reconstruct the evolution of baleen whales quite ambiguous. This is a typical problem for phylogenetic constructions of clades that diverged rapidly a long time ago.
In a recent study, Mark Springer and colleagues provide two new pipelines to study such hard to pinpoint diversification events: ASTRAL_BP (modified from ASTRAL; Mirarab & Warnow 2015) and SDPquartets (modified from SVDquartets; Chifman & Kubatko 2014). They developed these two pipelines to resolve species relationships using so-called retroelement insertions. But before I tell how they use retroelements, let me first describe the problems we face when we attempt to resolve these species relationships.
The problem
To study species relationships, we mainly use DNA sequences in the hope of finding mutations that are shared among subsets of species and that inform us about the order in which they split. However, at least three evolutionary principles complicate phylogenetic reconstructions of rapidly diversified species (Radiation; Figure 1).
First, at the time species split, they will share many mutations that were polymorphic in their common ancestral populations (Incomplete Lineage Sorting (ILS); Figure 1). It may take many thousands of generations after the species have gone their separate ways for these polymorphisms to uniquely sort among the species and unambiguously point out their correct relationships.
Second, while a goal in phylogeny is to find a point in time at which we can categorize independent evolutionary lineages, species do not always strictly abide to this ‘independence’ (Hybridization; Figure 1). Hybridization events can occur thousands of generations after the species split, whereby parts of the genome may swap and contain the evolutionary story of the other species (Edelman et al. 2019).
Third, as time goes on, mutations can occur at the same genetic position in separate species. As there are only four nucleotides to choose from (A, C, G and T), chances are that the two species independently acquire the same mutational variant. This will generate a phylogenetic signal as if they shared an ancestor more recently, a problem called homoplasy that increases for older speciation events (Figure 1).
Species relationships can be inferred despite Incomplete Lineage Sorting (ILS) and hybridization. This is done by not just investigating a single DNA sequence but by using and comparing multiple independent positions in the genome (in technical terms, this is done using a ‘multispecies coalescent’ model). In contrast, homoplasy is a harder phylogenetic problem that saturates phylogenetic signals, especially for groups of taxa that diverged a long time ago. This is where retroelements come in.
The solution
Retroelements or retrotransposons are a type of transposable elements (TEs) that copy and paste themselves by converting RNA back into DNA and inserting themselves into a new genomic location. In contrast to DNA sequence data,new retroelement insertions almost always occur at unique genomic locations and thus suffer very little to no homoplasy (Doronina et al. 2019).
As for mutations in DNA sequences, absence or presence of retroelements can be used to compile unique evolutionary fingerprints and reconstruct species relationships. Springer et al. (2020) do this by extending ILS-aware methods to be used with retroelements. With these new methods in hand, they reinvestigated the complex relationships of clades within the rorquals (Balaenopteridae), Placentalia, Laurasiatheria (bats, odd- and even-toed ungulates and carnivora) and Palaeognathae (one of the two living superorders of birds) and are able to confirm and resolve several conflicting relationships.
Springer et al. (2020) also provide a new method to test for ancient hybridization events. Their method is a Quartet-Asymmetry test that evaluates the frequencies of 4-taxon topologies similar to an ABBA-BABA test but not requiring an outgroup to be specified.
Interestingly, because retroelements are scored as absence and presence values, they also do not suffer from intralocus recombination, meaning that they can only represent one unique evolutionary history. This contrasts with long DNA fragments that may capture conflicting evolutionary events (for example, imagine a case in which half of a 20,000 bp DNA fragment has recombined with a sequence from another species after hybridization). Adding to the absence of homoplasy, retroelements thus also provide a powerful resource to study ancient hybridization.
For the baleen whales, Springer et al. (2020)’s inferred species relationships confirm those previously suggested from whole-genome sequencing, placing the gray whale as a sister lineage to the fin and humpback whale within the rorquals (Árnason et al. 2018).
Using their Quartet-Asymmetry test it becomes clear why gray whales have such hard to pinpoint phylogenetic position. Springer et al.(2020) confirm a multitude of hybridization events that involved the gray whales and resulted in conflicting phylogenetic signals (Figure 2).
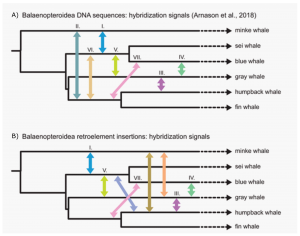
In conclusion, the use of retroelements and appropriate methods to analyze them provide a valuable asset to phylogenetics. As high-quality genomes are becoming more available for diverse groups of organisms, retroelements may become an important resource to reevaluate the relationships in a multitude of recalcitrant species groups (e.g. Arcilaet al.2017).
The study by Springer et al .(2020) is published in the Journal of Heredity with free access and comes with scripts for their newly developed tool SDPquartets and ASTRAL_BP.
References
Arcila D, Ortí G, Vari R, Armbruster JW, Stiassny MLJ, Ko KD, Sabaj MH, Lundberg J, Revell LJ & Betancur RR (2017) Genome-wide interrogation advances resolution of recalcitrant groups in the tree of life. Nature Ecology & Evolution 1: 0020.
Árnason Ú, Gullberg A & Janke A (2004) Mitogenomic analyses provide new insights into cetacean origin and evolution. Gene 333, 27–34.
Árnason Ú, Lammers F, Kumar V, Nilsson MA & Janke A (2018) Whole-genome sequencing of the blue whale and other rorquals finds signatures for introgressive gene flow. Science Advances 4: eaap9873.
Chifman J & Kubatko L (2014) Quartet inference from SNP data under the coalescent model. Bioinformatics30:3317–3324.
Doronina L, Reising O, Clawson H, Ray DA, Schmitz J. 2019. True homoplasy of retrotransposon insertions in primates.Systematic Biology 68:482–493.
Edelman NB, Frandsen P, Miyagi M, Clavijo BJ, Davey J, Dikow R, Accinelli GC, Van Belleghem SM, Patterson NJ, Neafsey DE, Challis RJ, Kumar S, Moreira G, Salazar C, Chouteau M, Counterman BA & Papa R, Blaxter M, Reed RD, Dasmahapatra K, Kronforst M, Joron M, Jiggins CD, McMillan WO, Di-Palma F, Blumberg AJ, Wakeley J, Jaffe D & Mallet J (2019) Genomic architecture and introgression shape a butterfly radiation. Science 366: 594–599.
Mirarab S & Warnow T (2015) ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics 31: i44–i52.
Nowak RM (1999) Walker’s Mammals of the World. Johns Hopkins Univ.Press, ed. 6.

Steven Van Belleghem is a Postdoctoral researcher at the University of Puerto Rico – Rio Piedras where he mainly focuses on studying color pattern diversity in Heliconiusbutterflies. Much of his work involves understanding the genomic architecture of phenotypic traits, studying population structure and contemplating speciation mechanisms. Before working on butterflies, Steven obtained his PhD at the University of Ghent in 2014 where he completed a project on the behavioral and genomic aspects of parallel and sympatric evolution in a ground beetle. You can learn more about his work here.




